GROUP DEBARBIEUX & STECHER / Associated Project
Mechanisms underlying bacteriophages and bacteria stable coexistence and its consequences on gut microbiome function
Bacteriophages are important effectors and indicators of human health and disease by managing specific bacterial population structures and by interacting with the mucosal immune system. Despite metagenome-based studies have addressed their abundance, diversity and stability over time in the gut, little in known on the role of bacteriophages in intestinal microbiota homeostasis and its impact on global microbiome functions. Furthermore, there is a gap in knowledge pertaining to the mechanisms by which bacteriophages and their bacterial hosts dynamically interact over time.
In this project, we will conduct an in-depth characterization of bacteriophage ecology and study their influence on the microbiome and related functions in the gut, using gnotobiotic mice colonized with a synthetic bacterial community, the Oligo-Mouse-Microbiota (OMM12). In addition, we will study the mechanisms underlying stable coexistence of bacteriophages and their host bacteria in the gut. The final goal is to refine strategies for bacteriophage-based intestinal microbiota engineering.
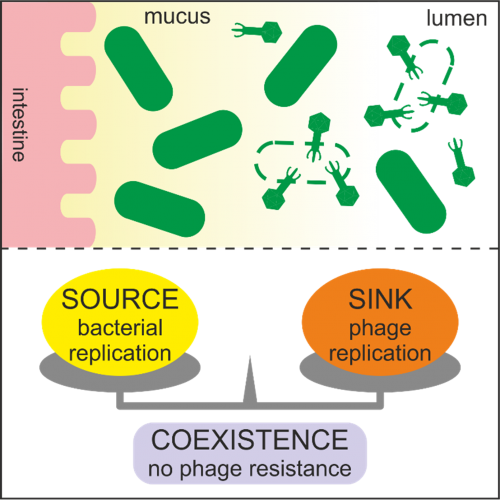
Figure 1: The uneven spatial distribution of bacteriophages in the gut creates bacterial refuges, which may hinder the overall efficacy of phage therapy targeting intestinal pathogens.
Principal Investigator(s)

Laurent Debarbieux
Bacteriophage, Bacterium, Host Laboratory
Institut Pasteur
E-Mail: laurent.debarbieux@pasteur.fr
Homepage: https://research.pasteur.fr/en/team/bacteriophage-bacterium-host/

Bärbel Stecher
Max von Pettenkofer Institute
LMU Munich
E-Mail: stecher@mvp.lmu.de
Homepage: http://www.mvp.uni-muenchen.de/en/research/medical-microbiology-and-hospital-epidemiology/group-stecher/
PhD student(s)
Devon Conti, devon.conti@pasteur.fr
Caroline Henrot, caroline.henrot@pasteur.fr
PostDoc
Luís Leónidas Cardoso, cardoso@mvp.lmu.de
Publications
Chromosome folding and prophage activation reveal specific genomic architecture for intestinal bacteria. Lamy-Besnier Q, Bignaud A, Garneau JR, Titecat M, Conti DE, Von Strempel A, Monot M, Stecher B, Koszul R, Debarbieux L, Marbouty M.Microbiome. 2023 May 19;11(1):111. doi: 10.1186/s40168-023-01541-x.PMID: 37208714
- Lamy-Besnier Q, Koszul R, Debarbieux L, Marbouty M. Closed and High-Quality Bacterial Genome Sequences of the Oligo-Mouse-Microbiota Community. Microbiol Resour Announc. 2021 Apr 29;10(17):e01396-20. doi: 10.1128/MRA.01396-20.
- Lourenço M, Chaffringeon L, Lamy-Besnier Q, Pédron T, Campagne P, Eberl C, Bérard M, Stecher B, Debarbieux L, De Sordi L. The Spatial Heterogeneity of the Gut Limits Predation and Fosters Coexistence of Bacteria and Bacteriophages. Cell Host Microbe. 2020 Sep 9;28(3):390-401.e5. doi: 10.1016/
- Spriewald S, Stadler E, Hense BA, Münch PC, McHardy AC, Weiss AS, Obeng N, Müller J, Stecher B. Evolutionary Stabilization of Cooperative Toxin Production through a Bacterium-Plasmid-Phage Interplay. mBio. 2020 Jul 21;11(4):e00912-20. doi: 10.1128/mBio.00912-20.
- Brugiroux S., Beutler M., Pfann C., Garzetti D., Ruscheweyh H.J., Ring D., Diehl M., Herp S., Lötscher Y., Hussain S., Bunk B., Pukall R., Huson D.H., Münch P.C., McHardy A.C., McCoy K.D., Macpherson A.J., Loy A., Clavel T., Berry D., Stecher B. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol. 2016 Nov 21;2:16215. doi: 10.1038/nmicrobiol.2016.215. PMID: 27869789.
- Nedialkova L.P., Sidstedt M., Koeppel M.B., Spriewald S., Ring D., Gerlach R.G., Bossi L. and Stecher B. Temperate phages promote colicin-dependent fitness of Salmonella enterica serovar Typhimurium. Environmental Microbiology. 2015 Oct 6. doi: 10.1111/1462-2920.13077.